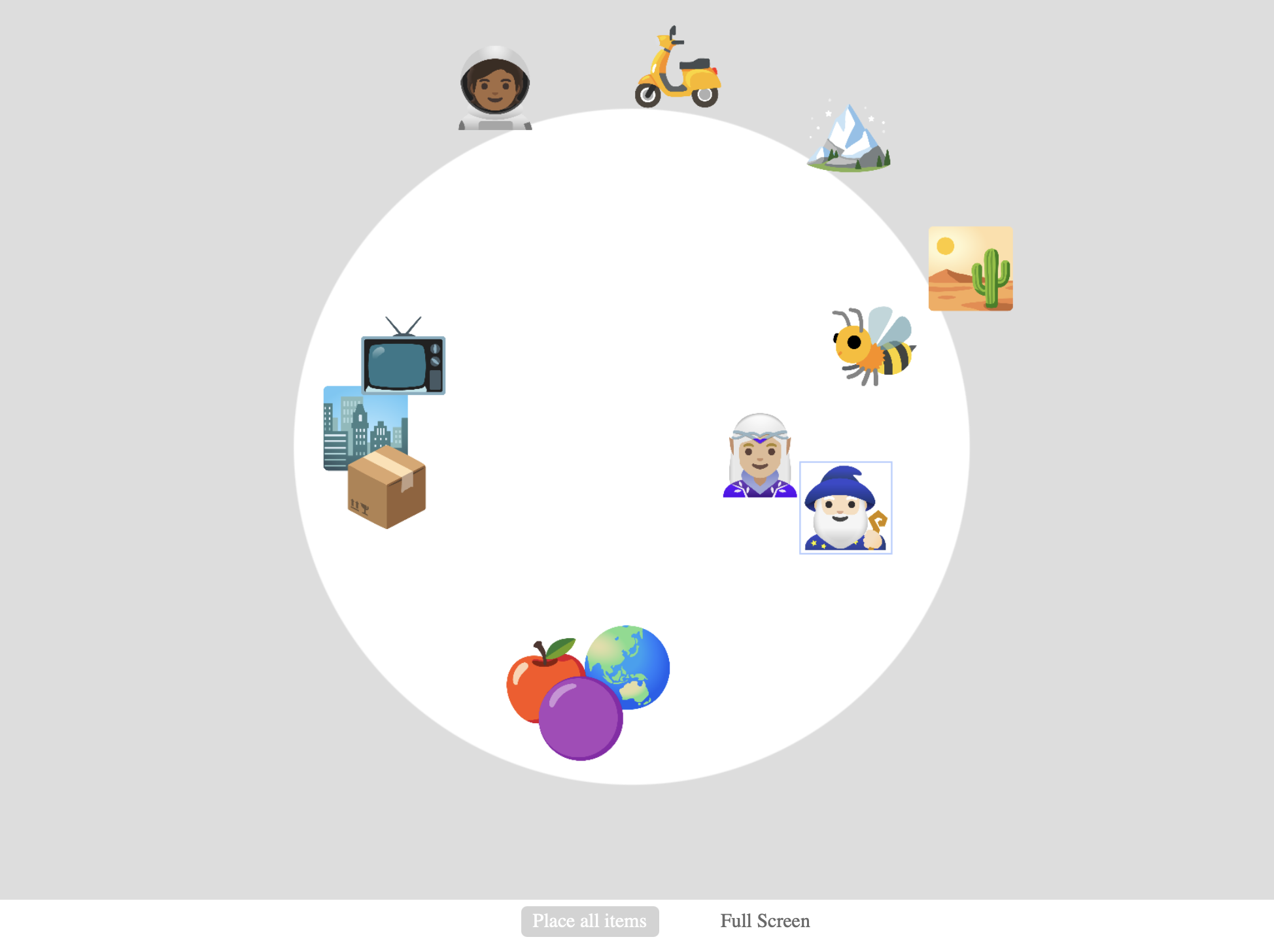
Multiple Arrangement Task¶
This is an implementation of Inverse MDS1, a high-dimensional innovation of the single-trial SPaM method. This paradigm is an efficient way to obtain subjective ratings of pairwise similarity among a large number of items.
In the task, the participant arranges three or more stimulus items in a circular arena, indicating by the distance amongst the items their pairwise dissimilarity. On Meadows, this can be done with image, text, video, and audio samples (where the items are represented by icons). To allow judgements on more than two dimensions, an initial arrangement trial is followed by arrangements of subsets.
Example Multiple Arrangement task, partially completed
The set of stimuli to be presented on the next trial are chosen with the lift-the-weakest algorithm1, a cost-benefit trade-off of participant effort versus additional evidence gained for the dissimilarity rating of each of the pairs involved.
Below you'll find information specific to the Multiple Arrangements task. This assumes you're familiar with how to setup an experiment and how to select stimuli for a given task.
Parameters¶
Customize the task by changing these on the Parameters tab of the task.
General Interface settings¶
Customize the instruction at the top of the page, as well as toolbar buttons. These apply to most task types on Meadows.
Instruction hint-
Text that you can display during the task at the top of the page.
Extended instruction-
A longer instruction that only appears if the participant hovers their mouse cursor over the hint.
Tip
As participants will typically be unfamiliar with this paradigm, we highly recommend using annotated images or video to instruct your participants.
Hint size-
Whether to display the instruction, or hide it, and what font size to use.
Fullscreen button-
Whether to display a button in the bottom toolbar that participants can use to switch fullscreen mode on and off.
Trials and stimuli¶
Control the number of trials, as well as the number of stimuli per trial.
Large stimulus sets: combining partial ratings across participants
One approach for larger stimulus sets (200+) is to obtain individual ratings for subsets and combine these into a larger, "fixed effects" or "group" dissimilarity matrix. Meadows has a built-in mechanism for efficiently subsampling the stimuli across participants, see sampling. You can use rsatoolbox to combine the partial RDMs .
Evidence utility exponent-
When determining which stimuli to select for the next trial, the evidence weight of each stimulus has this exponent applied to it, to calculate its utility if it were picked.
Min required evidence weight-
Among all pairs of stimuli, if the pair with the lowest evidence has an evidence weight higher than this, the task is done. The participant will be presented with new trials until this is achieved, or the time limit Max session length (see below) runs out, whichever comes first.
Max session length (in minutes)-
Total amount of time that the participant may spend on the task before the experiment continues. Time in between trials, or time spent on trials that are not submitted (i.e. the subject closes the browser) does not count towards this time limit. Default: 60 minutes.
Maximum number of objects presented in any one trial-
By default the first trial will involve all stimuli sampled for the task. If you set this number to a value that is smaller than that, a random subset will be used of that size.
Warning
With a maximum number of items per trial that is significantly lower than the total selection of stimuli, it may take many trials before a dissimilarity value has been obtained for every pair; this may result in missing values in the dissimilarity matrix if the task is finished (due to the time limit set) before every pair has been seen by the participant.
Tolerance-
How many stimuli can be left outside of the arena (unrated). These stimuli may re-occur on subsequent trials. By default all stimuli must be placed.
Spatial configuration¶
Options for customizing the lay out of the screen during the task.
Item Size-
Height of the stimuli in % of the width of the field. Default 8%. Must be between 2% and 20%. The width is adapted according to the original aspect ratio of the image.
Tip
To optimize signal-to-noise, and thereby task efficiency, instruct participants to use the full space of the circular arena; that is, place the pair of most dissimilar stimuli at opposite boundaries of the circle.
Stimuli start to the sides-
By default the stimuli start out surrounding the circle, this puts them to the right and left
Stimuli to the sides: Number of iterations-
Only applies if using "Stimuli start to the sides" above. Setting this higher than 1 means an algorithm will be run to find the best locations to minimize overlap. Depending on the number of stimuli this may be computationally intensive and therefore be disruptive for the participant. Use with caution!
Progress bar-
Display progress of alotted time at each trial
Magnifier-
Display the highlighted stimulus enlarged in the top-right corner
Magnifier size-
Size of a magnified stimulus. Same units as "item size"
Magnifier position-
Vertical position of the magnifier; either top or middle.
Arena position-
Vertical position of the circle. Currently only works for circular (not lateral) stimulus starting positions.
Mouse tracking-
Store mouse tracking data. This is a beta feature only unlocked on selected accounts. Contact us if interested.
Attention Checks¶
With these options you can enable attention check trials to be presented after
each regular arena trial. These are disabled by default and only appear if
attention checks per trial is set to more than zero.
This is an advanced feature and we are still determining the effectiveness.
Contact us to discuss details.
Attention check instruction heading-
This will be displayed on the screen during the attention checks phase.
Label for top pair-
The label displayed above the top pair in the attention check. The character $ will be replaced with the distance value.
Label for bottom pair-
The label displayed below the bottom pair in the attention check. The character $ will be replaced with the distance value.
Attention check prompt-
Optional custom question for the participant. Leave empty to use a (check type specific) default sentence.
Attention check type-
Which kind of attention check to use;
- confirm: Yes/no question on a textual representation of the distance.
- repeat: Yes/no question on a ranked representation of the distance.
- reproduce: Participant moves stimuli to reproduce the distance
- explain: Ask the participant for their reasoning.
Attention check scope-
Whether to present two ("pair") or three ("triplet") stimuli in a check trial. When presenting two, the distance is with respect to the maximal distance (the circle diameter). When presenting three, the distances of the pairs are relative to each other (which is more in line with the multiple arrangements concept)
Attention check sampling-
There are four different ways to determine the number of checks per trial:
- Fixed number per trial: every trial will have the same number of checks
- Percentage of Stimuli: the number depends on the amount of stimuli in this MA trial
- Percentage of Pairs: the number depends on the amount of pairs in this MA trial (non-linear)
These all refer to the number for Attention checks per trial
Attention checks per trial-
Number of checks per trial. For unit see Attion Check Sampling. The remainder will be treated stochastically. (E.g.
0.5means circa one check every other trial) max attention checks per trial-
Upper limit to the number of attention checks in any given trial. Used to limit the number from
Attention checks per trial. Random distance exponent-
A higher value makes small distances more common.
Distance unit-
How to describe distances; either as a fraction or a percentage.
Data¶
For general information about the various structures and file formats that you can download for your data see Downloads.
As part of a 1D or 2D "Matrix" in a matlab file:
rdmutvThe condensed or "upper triangular vector" form of the pairwise euclidean distances.stimuliThe stimuli of the task, dictating the order forrdmutv
As stimulus-wise "annotations" (table rows), with columns:
trialnumerical index of the trialstim1_idmeadows internal id of the stimulusstim1_namefilename of the stimulus as uploadedxrelative horizontal coordinateyrelative vertical coordinate
In the Tree structure:
rdmAn array containing the upper triangular vector of pairwise dissimilarities.next_trial_stimuliAn array with the IDs of the stimuli that would be presented to the participant next.trialsAn array with a key/value map for each of the trials that the participant finished:startTimestamp or epoch time of the start of the trial, in seconds since 1/1/1970.endTimestamp epoch time of the end of the trial, in seconds since 1/1/1970.- positions An array with a key/value map for each of the stimuli:
xrelative horizontal coordinate of the stimulus.yrelative vertical coordinate of the stimulus.idThe ID of the stimulus, corresponding to the ID field in the stimulus list.
Analysis and Visualization¶
We recommend the python library rsatoolbox for performing analysis on similarity ratings.
Plot ratings as representational similarity matrix¶
import json
import numpy
from scipy.spatial.distance import squareform
from matplotlib.pyplot import matshow, show
# read "tree" data for a single participant
file_path = 'my_data/Meadows_myExperiment_v1_tree.json'
with open(file_path) as fhandle:
data = json.load(fhandle)
# Assumes the MA task is the first (index 0)
upper_triangular_vector = data['tasks'][0]['rdmutv']
# Convert the condensed vector to a matrix
rdm = squareform(upper_triangular_vector)
# display the matrix
matshow(rdm)
show()
% read "1D array" data for a single participant
file_path = 'my_data/Meadows_myExperiment_v1_1D.mat'
data = load(file_path)
% Assumes the MA task is the first (index 0)
upper_triangular_vector = data['tasks'][0]['rdmutv']
% Convert the condensed vector to a matrix
rdm = squareform(upper_triangular_vector)
% display the matrix
matshow(rdm)
References¶
-
Kriegeskorte, N., & Mur, M. (2012). Inverse MDS: Inferring dissimilarity structure from multiple item arrangements. Frontiers in Psychology, 3, 245. doi: 10.3389/fpsyg.2012.00245 ↩↩